Single-Molecule Peptide Identification using Fluorescence Blinking Fingerprints
The analysis of the proteome is complicated by the presence of isoforms, post-translational modification (PTM), and the insufficient correlation between the abundance of protein and the transcriptomic or genomic information. As the current state-of-the-art, mass spectrometry-based proteomics methods remain limited in their sensitivity and dynamic range compared to the established single-molecule approaches in genomics and transcriptomics.[1,2] In particular, single-molecule identification of peptides and proteins would enable the analysis of biomarkers that are present in very small quantities, for example in diluted clinical samples, single cells, or isolated organelles.[3]
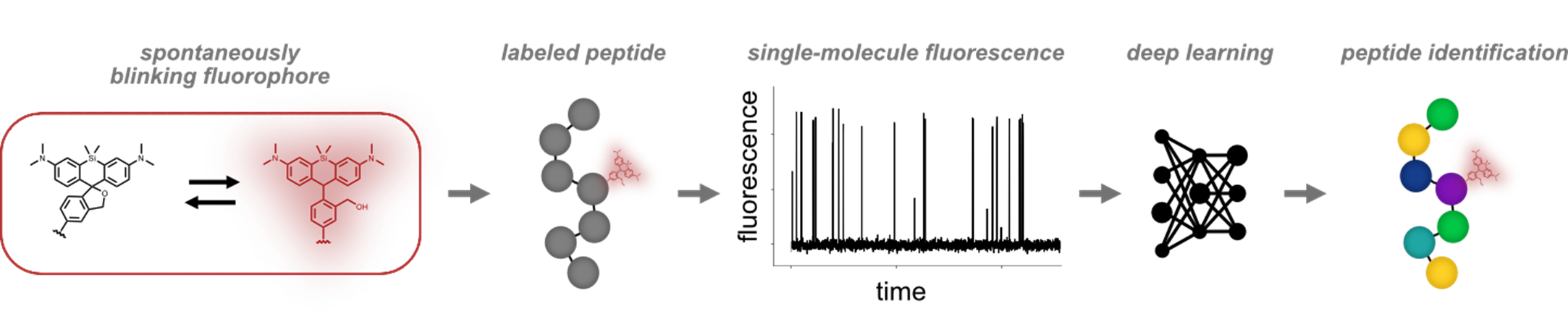
In this work, we provide a proof-of-concept of a fundamentally new approach to identify single peptide molecules, including subtle PTMs, that does not rely on sequencing.[4] Although this method is presently constrained to targeted studies, it holds potential as a widely applicable, rapid, and accurate single-molecule proteomics technology. Our approach exploits the emission of a spontaneously blinking fluorophore to capture information on the chemical environment of the probe.[5] We use single-molecule fluorescence measurements and a deep learning model based on convolutional, gated-recurrent unit layers to identify the peptide of interest in a targeted manner. By implementing Monte Carlo dropout, we obtain an uncertainty measure for classification which we use for filtering out low-quality traces. We first validate our method on a small set of unnatural peptides of the same length but varying amino acid sequences, which allows us to classify single peptide molecules with overall accuracies > 90 %.[4] Furthermore, the method can be applied to differentiate between peptides with a variable number and position of PTMs. The applicability of the analysis is demonstrated for two PTMs, including the epimerization of an amino acid which would be very difficult to analyze by mass spectrometry. We envision that the technology is adaptable to molecules beyond peptides and proteins depending on available conjugation strategies. Moreover, the method has the potential for further refinement in experimental and analysis aspects to improve the accuracy and extend the applicability to real biological samples.
[2] Bruno Domon, Ruedi Aebersold, Nat. Biotechnol., 2010, 28, 710-721.
[3] Javier Antonio Alfaro, Peggy Bohländer, Mingjie Dai et al., Nat. Methods, 2021, 18, 604-617.
[4] Salome Püntener, Pablo Rivera-Fuentes, J. Am. Chem. Soc., 2023, 145, 1441-1447.
[5] Shin-Nosuke Uno, Mako Kamiya, Toshitada Yoshihara et al., Nat. Chem., 2014, 6, 681-689.