The conservation of protein dynamics on an evolutionary timescale of nearly a billion years
Proteins are the fundamental building bricks for living organisms. Their structure, dynamics, and function are inherently encoded in their amino acid sequence. In the course of evolution, the conservation of protein sequence, structure, and dynamics is depending on selective constraints and differs from protein to protein [1]. Although the conservation of protein sequence and structure has been extensively investigated in the past, very little is known about the conservation of protein dynamics. Does the conservation in protein structure correlate with the conservation of protein dynamics?
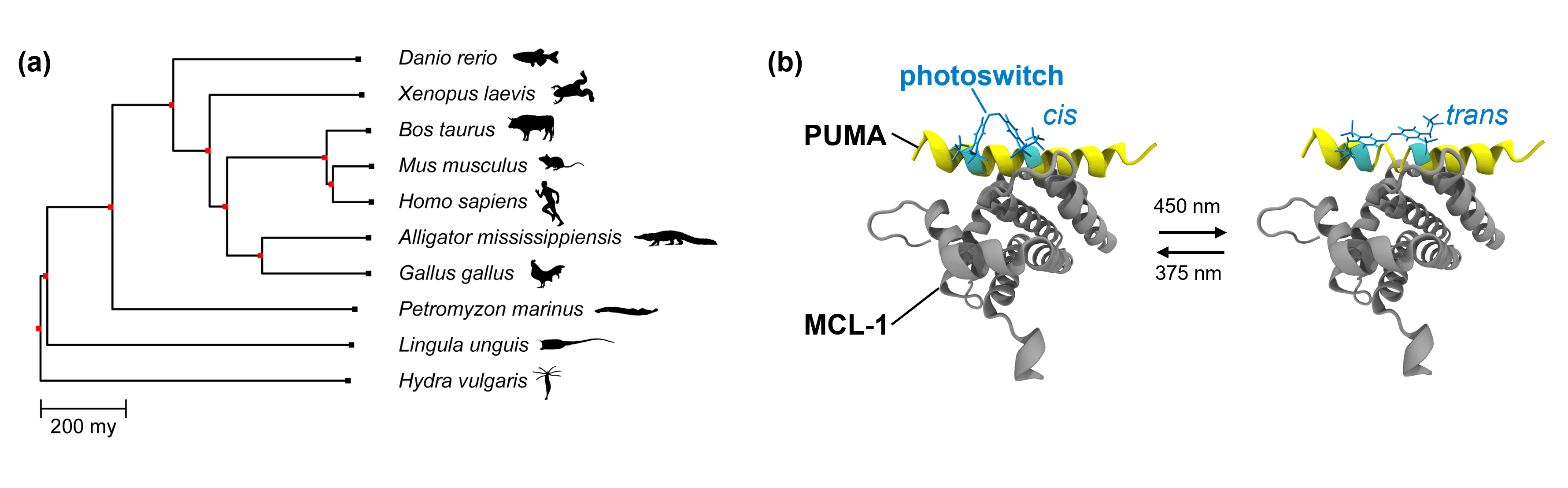
In our current study, we investigate the dynamics of the protein MCL-1 from ten different species, separated by nearly a billion years of evolution (Figure 1a), via transient infrared spectroscopy. MCL‑1 is a key regulator in apoptosis, the programmed cell death [2]. Due to its crucial function in apoptosis, MCL-1’s structure and function appears to be conserved in all vertebrates and a lot of other animal species. Here, we generated photoswitchable versions of MCL-1 in complex with its natural binding partner PUMA (Figure 1b) to investigate the dynamic response of the protein complex upon photo-induced perturbation [3]. When triggering the photoswitching event in PUMA, we altered PUMA’s secondary structure in the binding pocket of MCL-1 and recorded the time scales of molecular response in a time window of pico- to microseconds.
By experimentally determining the dynamic behavior of these ten photo-perturbed MCL-1 homologs, hundreds of million years apart, we detected similar patterns. We found dynamic processes in the nanosecond regime which correlate well with the evolutionary separation of the protein homologs. Our findings help to understand how fast molecular processes change over an evolutionary timescale and which kind of dynamic processes remain preserved.
[2] J. Kale, E. J. Osterlund, D. W. Andrews, Cell Death and Differentiation 25, 65 (2018).
[3] P. J. Heckmeier, J. Ruf, B. G. Janković, P. Hamm, Journal of Chemical Physics 158, 095101 (2023).